Mass spectrometry is an analyticaltechnique that can be used to study the compositional make-up of complex biochemical samples with high specificity. This technology has become the gold standard in life science fields such as proteomics, metabolomics, and toxicology. This is due to its specificity and ability to quantify miniscule amounts of hormones, diseases, biomarkers, metabolites, and more.
Challenges Facing Mass Spectrometry
Although mass spectrometry has been widely adopted, there remain considerable challenges that impede its adoption in clinical and laboratory settings.
The most obvious barriers include low analytical throughput for routine analysis of large, isomeric samples and low spatial resolution, where measuring within the anatomical context of dissociated tissues on the micron to millimeter scale holds great significance.
Mass Spectrometry Imaging (MSI)
Imaging mass spectrometry (IMS) is a highly sensitive molecular imaging technique that provides combined molecular information and spatial resolution. It enables the direct identification and localization of compound molecules from tissue sections, single cells, or other surfaces. Core features of IMS research are the high sensitivity of mass spectrometers, the untargeted nature of the technology, imaging capabilities for peptides and proteins, and spatial resolutions from the organism level (hundreds of microns) down to the cellular level (tens of nanometers). IMS allows simultaneous imaging of thousands of different molecules in a single experiment.
Thus, it is an effective multi-component molecular imaging technology. Scientists have developed many different IMS workflows and instruments to study endogenous biological compounds like lipids, peptides, and proteins, as well as exogenous compounds like polymers, or to study tissue distributions of administered drugs. These studies provide intricate details of biological processes from the subcellular to organism levels.
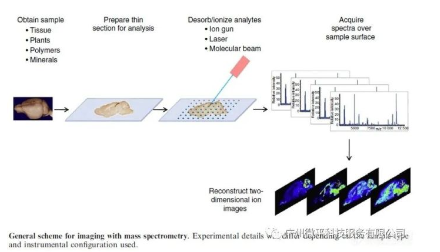
Today, imaging mass spectrometry is mainly a technique for pathological studies of dissociated tissue sections, and does not have the capabilities for in vivo diagnostics like MRI or PET scanning. However, it can serve as a complementary technology for in vivo imaging to verify distributions and metabolic fates of biomolecules across disease stages or modes of drug delivery. Many researchers are exploring how this complementary imaging approach can address specific questions about molecular distributions. The rationale for this approach is clear. No other single imaging technology provides an adequate combination of molecular structure and anatomical information with proper spatial, temporal, and biological state resolution.
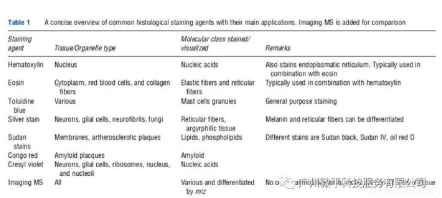
Compared to other molecular imaging methods like MRI, PET, or immunohistochemistry (IHC), imaging mass spectrometry (IMS) has a unique capability to visualize compound molecules without the need for labels, enabling exploratory research into distribution patterns of novel compounds not feasible with the other technologies. It is often the only tool capable of identifying compound molecules when using conventional histological stains that impact chromaticity, such as hematoxylin and eosin (H&E) commonly used for tissue staining.
It can be used to study compound molecular distributions not feasible with routine histological stains. This is because the conventional stains used in pathology provide only general histotypings, and do not identify specific molecules, provide molecular modification details, or combinatorial information. Some examples of drugs and metabolites that cannot be stained by common histological stains are listed in Table 1.
Ionization and Desorption Techniques for MSI
IMS requires desorption and ionization of compound molecules from the surface of the specimen being studied. There are two main physical approaches: (1) collision of the analytic surface with energetic charged particles, (2) irradiation of the surface with photons from a pulsed coherent light source.
1. Desorption and Ionization by Charged Particles
Charged particles are mainly used for secondary ion mass spectrometry (SIMS) imaging. In this approach, the analytic surface is exposed to a focused high energy primary ion beam. Ion impact causes a cascade of collisions of molecules up and down from the surface, leading to ejection and ionization of surface molecules.
The collisionally generated secondary ions can then enter a mass analyzer to determine their nature. The collision energy is usually kept low to ensure the primary ions can interact with surface molecules in different areas, and to ensure the collided areas are not analyzed again in secondary collisions. Experiments with collision energies below the surface monolayer analysis threshold are termed static SIMS experiments. Experiments above that collision energy are termed dynamic SIMS experiments. The analytic surface undergoes continual change during dynamic SIMS experiments. In static SIMS experiments, typically less than 1% of the analyzed surface is consumed.
A large amount of internal energy is transferred to the surface molecules during SIMS experiments. This causes extensive fragmentation of compound molecules in the surface layer. Thus the method is less suitable for direct analysis of large biomolecules like peptides and proteins. The method works better for observing distributions of surface elements and small molecule compounds. Fragmentation patterns are similar to those observed with electron impact ionization.
The most commonly used primary ion species are indium and gallium. They are mainly applied to the analysis of elements and organic impurities on semiconductor surfaces, and studies of surface coatings on thin layers. Benefiting from the application of larger cluster ions or molecular ions, biological surfaces like tissue sections can also be analyzed. Larger primary ions like Aun+, Binm+, and C60+ are examples. These ions can yield higher intact secondary molecular ion yields, and reduce fragmentation of molecular ions. Additionally, use of these ions can significantly reduce damage to subsurface molecules, increasing the chances of success for 3D imaging experiments.
All SIMS experiments require a vacuum environment with the ion beams described above, otherwise the mean free path of primary ions would be too short for them to reach the analytic surface. Desorption electrospray ionization (DESI) is a desorption and ionization technique at atmospheric pressure. It produces electrospray droplets that are then transported to the analytic surface under ambient conditions.
The solvent droplets extract molecules from the surface, leading to secondary ion generation similar to conventional electrospray mass spec ionization. This approach can produce multiply charged quasi-molecular ions. It has been reported suitable for surface analysis of a variety of specimens including pharmaceutical tablets, blood stains, and tissue sections. Studies have shown DESI technology can be used for tissue imaging to visualize distributions of phospholipids and lipids in brain and tumor tissue sections.
2、Photon desorption, ionization
(1) LDI and MALDI
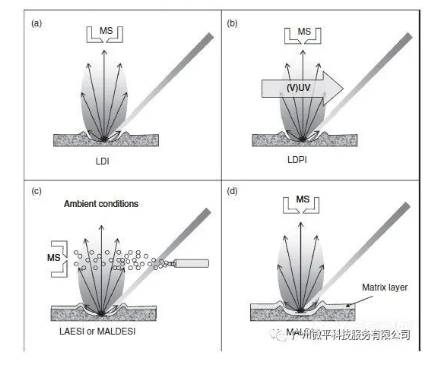
A second way photons can desorb and ionize molecules from a surface is via their interactions with surface molecules. Typically a pulsed laser beam is focused on the analytic surface. The photon energy absorbed by the surface layer causes explosive ejection or ablation of the surface material. When using infrared (IR) or visible light, the photon energy is mainly converted into vibrational energy in the surface. Under ultraviolet or vacuum ultraviolet (VUV) light, the increased photon energy can lead to extensive electronic excitation. If the internal energy accumulated in the analyte molecules is sufficient to cause direct ionization, the process is termed laser desorption ionization (LDI), as shown in Figure 1(a). The internal energy accumulated during laser desorption is typically quite high, and can cause extensive fragmentation of surface molecules. Additionally, the low ionization efficiency of organic compounds makes the technique less suitable for macromolecular mass spectrometry.
In these cases, laser desorption post-ionization (LDPI) strategies can be applied to ionize the neutral particles generated in the desorption process (Figure 1(b)). Post-ionization strategies can be achieved under vacuum conditions by irradiating the desorbed plume with secondary energy laser beams in the UV or VUV wavelength range. Recent studies show laser desorption can be effectively coupled with ESI ion sources, enabling laser ablation electrospray ionization (LAESI) under atmospheric pressure conditions (Figure 1(c)). This combination expands the compound classes that can be analyzed by laser desorption strategies, and can reduce compound fragmentation. When combined with inductively coupled plasma mass spectrometry (ICP-MS), laser ablation can be successfully used for quantitative analysis of surface elements in a sample. The ablated components are atomized and ionized into elemental and isotopic ions by the ICP source, which are then analyzed by the mass spectrometer. Additional quantitative information can be obtained from the light emitted by the ICP using optical emission spectroscopy.
Due to extensive fragmentation, direct LDI strategies are not suitable for analysis of biomolecules larger than 500 Da. In these cases, energy-absorbing matrixes can be used. Mixing or coating the analyte on the surface to be analyzed (see Figure 1(d)) can overcome this limitation. This technique, conceived by Karas and Hillenkamp in the late 1980s, is termed matrix-assisted laser desorption ionization (MALDI). It is a key technique in modern proteomics research, and can be applied to desorption and ionization of large biomolecules like proteins and DNA. In MALDI analysis of complex sample surfaces, matrix-assisted approaches have greater utility.
First, application of a matrix can reconstitute the analyte molecules from a complex sample within or on the surface of matrix crystals. The formation of these analyte-doped matrix crystals can separate the analytes from other assisting factors like salts, and can disperse large molecules within the matrix. Subsequent pulsed laser irradiation of the crystal surface can rapidly superheat the sample. This is a direct consequence of electronic excitation (UV-MALDI) or vibrational excitation (IR-MALDI) of the matrix acting as a strong absorber of the laser energy. The synergistic desorption of the superheated matrix carrying the analytes can be directed into the vacuum. This facilitates a non-destructive transition of the analyte molecules into the gas phase. The final role of the matrix is to promote ionization of the analyte molecules via charge transfer. This typically yields [M+X]+ type cations as intact quasi-molecular ions, where X denotes the type of cation generated. The most common cations are hydrogen, sodium and potassium. For analytical success, the analyte molecules must cocrystallize with the solid matrix material, and this matrix should be in excess. Typical matrix-to-analyte ratios are in the range 103:1 to 105:1. Empirically, the higher the mass of the analyte under study, the more excess matrix is required for complete desorption.
(2) Applications of MALDI in open environments
Recent developments in open desorption strategies, which also utilize matrices, have made some advances. Similar to LAESI methods, their matrix-analyte mixtures need to cocrystallize on a substrate, allowing more intact sample to be removed from the surface. MALDI ions experience deflection by the electric field between the mass spectrometer inlet and sample surface. The neutrals generated from MALDI matrices contain a significant amount of analyte molecules lost in vacuum MALDI experiments. These can be adsorbed onto the surface of incompletely atomized electrospray droplets. What follows is the conventional generation of multiply charged ions by electrospray ionization. Abbreviated as MALDESI (matrix-assisted laser desorption electrospray ionization), it can combine the advantages of MALDI in open environments and the sensitivity of electrospray ionization.
(3) MALDI and liquid chromatography
The successful coupling of MALDI techniques and liquid chromatographic (LC) separation techniques improves the separation and detection efficiency of complex mixtures. MALDI experiences significant ion suppression when analyzing complex mixtures. The coexistence of compounds with differing physiochemical properties often leads to preferential ionization of one or a few components over others. Ion suppression effects are a major impediment to quantitative studies across many analytical disciplines. Explanations for MALDI mass spectral intensity discrepancies are inherently qualitative in nature. One approach to overcome this is chromatographic separation to reduce the complexity of the mixture. Many nano-LC-MALDI methods have been implemented that convert the separation time scale into a spatial distribution scale. Automated spotting techniques can deposit a series of 2 nL liquid chromatographic eluent droplets (typically 150 nL each) onto a pre-coated MALDI matrix surface. Alternatively, matrix solution can be mixed with LC eluent and this mixture deposited as ordered droplets on a clean matrix target for mass analysis.
3. Use of matrices in SIMS
The advantages of using energy absorbing matrix materials are not limited solely to photon desorption and ionization techniques. The success of MALDI mass spectrometry made possible the application of MALDI matrices in SIMS (secondary ion mass spectrometry) sample preparation. Cocrystallization of the analyte with a MALDI matrix (2,5-dihydroxybenzoic acid/DHB) facilitated detection of large molecular ion species >10 kDa by matrix enhanced SIMS (ME-SIMS). Thus, this technology to produce intact large molecular ions (peptides, proteins, oligonucleotides) solely based on SIMS ionization principles was successful.
It has been proposed that the role of the matrix in ME-SIMS is similar to that in MALDI: providing a nestled environment for the analyte molecules and supplying protons to enhance ionization. DHB gives the best results as matrix, possibly explained by DHB increasing the concentration of analyte in the near-surface regions of the sample. Since ME-SIMS probes only the top 50 nm (compared to MALDI), positioning of the analyte molecules is critical for sample preparation. The analyte molecules must be present at the surface of the crystals because deeper layers of the matrix cocrystals cannot be detected under the static SIMS conditions.
Spatial Resolution in Imaging Mass Spectrometry
A key parameter in IMS is the achievable spatial resolution. The spatial resolution determines the observable details on the surfaces of cells and tissues. The most common approach to obtain mass resolved images is by using microprobe or rastering modes. Microprobe mode mass spectrometry imaging scans an ionizing probe beam across the sample surface in SIMS or moves the sample through a focused MALDI irradiation. For each discrete location, interaction of the ionizing beam with the sample stores coordinates and obtains position correlated mass spectral data from generated ions. In this way a raster is constructed where each raster point has an associated mass spectrum.
Using dedicated software, ion images mass resolved can be constructed from these datasets. The maximum achievable spatial resolution in microprobe imaging experiments is determined by the size of the microprobe. Technically, the precision of rastering each point in the grid is another factor controlling resolution but this is usually not an issue for SIMS and MALDI imaging. Additionally, the experimentally realized spatial resolution is influenced by factors related to sample preparation (matrix) and sensitivity (signal-to-noise).
1. Spatial resolution of SIMS and DESI imaging mass spectrometry
SIMS mostly uses ion sources constructed from liquid metal ion guns. Ga+ and In+ are mainly used for surface elemental and small molecule analysis. Spatial resolutions down to 50 nm can be achieved with these guns as determined by the emitter size, electrostatic optics in the ion column and the primary beam current. The latter is often kept low to prevent spatial charge spreading of the beam and loss of resolution. When tuned at low currents, both guns can provide a 50 nm focus. Metal cluster beams like Aun+, Bin+ and C60+ can provide spot sizes of 100-200 nm at very low beam currents. The low beam currents often require longer experiment times. Thus, to apply higher beam currents to increase analysis speed, the spatial resolution is commonly compromised and reduced to around 1 μm.
DESI uses a charged solvent droplet spray directed at the surface. The size of the wetted interaction area on the surface determines the spatial resolution. Studies show DESI imaging routinely achieves ca. 1 mm spatial resolution.
2. Spatial resolution of laser desorption ionization (LDI) and matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry
The resolution of a focused laser beam is wavelength determined and limited by the Abbe diffraction limit. Long wavelength infrared lasers struggle to focus below 50 μm. The physical spot sizes of UV lasers in commercial instruments are restricted to around 10 μm. In commercial instruments, most experiments use laser spot sizes between 50 and 250 μm. This choice is dictated by sensitivity and time required to complete experiments. Specialized confocal objectives can reduce spot sizes down to 1 μm but there remains substantive debate if using MALDI these small spots require laser threshold fluences too high for non-destructive analysis of compounds in tissues.
Preliminary experiments have shown its capabilities for acquiring high resolution images of analytes. An alternative approach to increase spatial resolution with conventional MALDI-ToF instruments is an oversampling method. In this method, the movement increment of the laser probe is smaller than the light spot diameter. All sample after the first sampling point is interrogated for information from areas smaller than the laser focus size with each sampling increment, thereby accomplishing increased spatial resolution. Two drawbacks of this method are limited tandem MS possibilities and greater total sample consumption.
Developments and Improvements
Using the desorption and ionization techniques described in the previous section, atomic and molecular ions can be generated from complex surfaces. Generation of mass spectrometric images requires subsequent mass analysis of these generated ions. Modern mass spectrometry provides a range of mass analyzers to accomplish this. This article introduces three types of mass analyzers that provide unique analytical capabilities for MALDI or SIMS mass spectral imaging of biological surfaces.
1. Time-of-flight mass spectrometry (TOF-MS) is the most commonly used mass analyzer in ion mobility spectrometry (IMS). It requires pulsed ions, a requirement that is ideally compatible with MALDI and SIMS. All ions are given the same accelerating potential. Ions of the same m/z will gain the same kinetic energy in addition to their initial kinetic energy imparted during desorption. Thus, their velocities depend on their m/z and ions can be separated by drifting in a field-free region. Ion detection is accomplished by a particle detector of the multi-channel plate (MCP) type. TOF analysis provides a very wide mass range, limited only by the detection sensitivity for high molecular weight species. MALDI-TOF-MS can analyze molecules up to several hundred thousand Daltons. The high duty cycle and total flight times in the microsecond range provide the possibility to use high repetition rate lasers for sensitive surface detection. This enables high-throughput analysis, a key requirement for analysis of large area samples. Improvements in resolution can be achieved by compensating for the initial kinetic energy imparted in the desorption process. Using delayed extraction, hemispherical electrostatic lens elements and reflectrons, mass resolving powers (FWHM) of m/Δm = 30,000 at m/z 1000 can be achieved. Tandem mass spectrometry for compound identification is typically accomplished by collision-induced dissociation (CID) or by observing the decay of metastable ions after ionization. For this, two independent TOF systems can be coupled in a so-called TOF/TOF configuration. The first TOF is used for precursor selection, the second TOF for product ion analysis.
2. Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) is an ion trapping technique that determines the cyclotron frequencies of ions trapped in a Penning ion trap in a strong magnetic field. After generation of ions in an external ion source, ions are transferred into the Penning trap until further analysis. Using broadband radiofrequency excitation, all ions are excited into large cyclotron orbits. Not only does their orbital radius increase, but ions of the same m/z also coherently orbit in the trap. During the orbit, they can induce oscillating image charges in a pair of detection electrodes. This time domain signal is digitized and Fourier transformed to produce a cyclotron frequency spectrum. The mass spectrum can be calibrated by using the cyclotron equation w=qB/m. The main advantages of FT-ICR-MS are unmatched mass resolution and mass measurement accuracy, which can be used to discover new structural details from MALDI imaging. In addition, the use of ion trapping techniques allows not only CID but also infrared multiphoton dissociation (IRMPD) and electron capture dissociation for tandem MS structural determinations. Analysis speed is limited by the length of the time domain signal observed and the desired mass resolution. Mass resolution depends on the coherent orbital time of the ions. Typical analysis times are 1 s per pixel, independent of the ionization source used. Shorter transient lengths for the same resolution can be achieved by increasing the magnetic field strength. MALDI tissue imaging experiments can be performed on FT-ICR-MS systems with FWHM resolution ranging from 40,000 to 400,000.
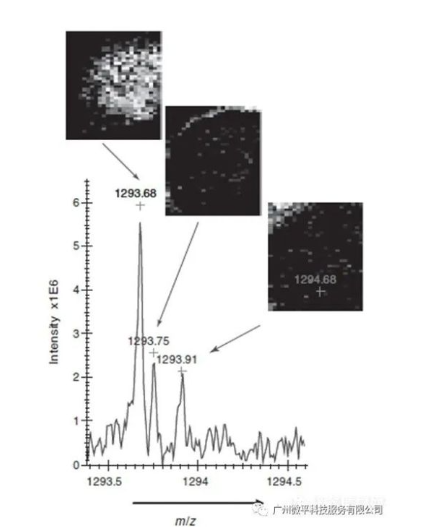
3. Matrix-assisted laser desorption/ionization ion mobility imaging mass spectrometry (MALDI-IM-IMS)By incorporating ion mobility separation of the ions generated by MALDI, additional information can be obtained in the mass spectra. Ion mobility spectrometry is a separation technique based on the collision cross-sectional area of ion migration. In ion mobility mass spectrometry, a gas-filled drift tube is used for ion separation prior to mass spectrometric analysis. These ions have different collision cross-sections due to conformational or compositional changes. When used for mass spectrometric imaging, in addition to the spatial and mass spectral dimensions, an extra dimension of gas-phase separation by drift time is added. Ion mobility spectrometry benefits MALDI imaging mass spectrometry research in two major aspects. First, the additional separation dimension enables detection of more spectral peaks. Ion mobility facilitates reducing the complexity of mass spectrometric analysis and aids in the separation of different classes of compounds, such as peptides and phospholipids. Second, the combination of mass and drift time selection allows the fragmentation of isobaric peptides or other analogs into product ion spectra. The combination of ion mobility, MALDI, and ToF-MS for IMS can localize and identify proteins through their associated proteolytic peptide fragments. Ion mobility separation can identify isobaric ions that cannot be identified by conventional MALDI-ToF-MS. Compared to traditional MALDI-ToF, this method increases the number of peaks observed per measurement, generates ion images with mass and time selections, and can identify individual ions.
The results shown in Figure 3 demonstrate the feasibility of protein identification from tissue by ion mobility flight time imaging mass spectrometry (IM-ToF-IMS).
Tissue digestion combined with the MALDI-IM-ToF-IMS method allows a "bottom-up" strategy for the identification of different tissue proteins.
MALDI Imaging Strategies
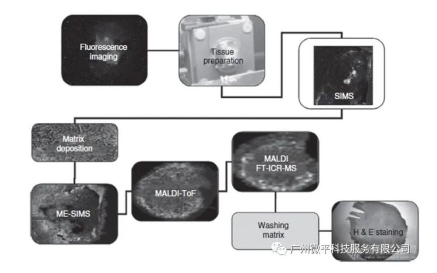
1. The combination of different desorption/ionization methods and mass analyzers in mass spectrometry imaging provides the possibility to perform complementary experiments on a single tissue sample. Careful experimental design is needed to ensure relevant complementary molecular image information is obtained. The experimental workflow shown in the figure provides an example of generating six complementary image datasets from a single tissue sample. In this example, a piece of tissue is obtained surgically. Cells within the tissue express a fluorescently labeled protein, so the first step in the imaging workflow is to generate a fluorescence image. This provides detailed localization of a specific protein. After sectioning 10-20 μm thick slices on the substrate surface and mounting, SIMS analysis is performed. This provides low molecular weight imaging MS data at high spatial resolution. Static SIMS removes less than 1% of the surface material, so the remaining surface can be further analyzed. After the SIMS study, the tissue surface can be coated with a matrix (see Matrix Coating section). Depending on the analytes of interest, the surface can or cannot be washed before matrix application. The wash protocol has an important influence on the results obtained. In the experimental workflow in Figure 4, no washing is done prior to matrix deposition to allow imaging of small water-soluble molecules. After matrix deposition, the first analysis performed is ME-SIMS. Again, only a small fraction of compound molecules are removed from the surface, leaving the crystalline surface intact for subsequent MALDI analysis. The ME-SIMS dataset provides information on larger intact organic molecules (such as lipids and small signaling molecules <2000 Da). The next analysis performed is MALDI-ToF analysis with a laser fluence slightly above the desorption threshold. The MALDI-ToF dataset contains information on endogenous peptides and intact proteins (depending on wash protocol and matrix used). The final MS imaging dataset that can be obtained is a MALDI-FTICR-MS dataset (or ion mobility image dataset). These techniques require removal of most of the matrix material. They can provide high mass resolution and mass accuracy information to aid identification of molecules constituting the images. Any remaining matrix material can be washed from the multiply analyzed surface to enable final H&E staining. This provides additional histological information that can be combined with the imaging MS datasets to identify specific regions or tissue types.
2. Matrix Coating
Prior to MALDI and ME-SIMS analysis, a matrix solution must be applied to the tissue surface. Matrix solutions consist of organic solvents like methanol or acetonitrile with added weak organic acids such as sinapinic acid (SA) or 2,5-dihydroxybenzoic acid (DHB) and trifluoroacetic acid (TFA). Addition of TFA increases the protonation of molecules. The method of matrix application will strongly influence imaging MS results. The application method will impact sensitivity, surface diffusion and spatial integrity, spatial resolution, surface flatness, and analysis speed. Tissue properties and environmental parameters influence protein extraction efficiency from tissue and matrix crystallization. Thus, controlling the matrix deposition environment is also very important. Several labs are considering innovative deposition methods such as matrix sublimation. For general labs, there are typically two matrix deposition methods: spotting and spraying.
(1) Matrix Spotting
When spotting matrix solution onto portions of tissue, diffusion of analytes needs to be constrained within the spot size. Two matrix deposition approaches have been developed: manual or automated spotting. Manual spotting produces droplets which are often used for non-imaging MALDI tissue analysis. Automated spotting utilizes smaller volume (pl) droplets and produces spot sizes of ~120-150 μm and minimum resolutions of ~200 μm. Two different types of automated spotters are used for matrix deposition: piezoelectric inkjet nozzles and droplet dispensers using focused acoustic waves. Both ejectors can dispense 100 pl droplets on tissue which dry to 150 μm diameter spots. In this case, the resolution of imaging MS analysis is often limited by matrix spot sizes larger than the analysis beam diameter.
(2) Matrix Spraying
Matrix spraying covers the entire surface of the sample with a uniform mist of small matrix solution droplets. Pneumatic, vibrating nozzles, or electrospray can generate the matrix solution spray. Spraying can be done manually or in an automated fashion. Manual spraying utilizes a hand-held airbrush or TLC sprayer. Automated spray application can be achieved by combining spray devices with an x-y robot, and allows matrix deposition over larger areas. Automated spraying over smaller areas can also be achieved using vibrating sprayers, where a small chamber mainly controls humidity. Resulting crystal sizes after spraying are typically 10-20 μm. To obtain smaller crystals, electrospray can be used which reduces sensitivity but produces crystals even smaller than 1 μm. When spray deposition is used, the diameter of the laser beam limits the spatial resolution of MALDI imaging mass spectrometry.
3. Identification Strategies
The identification of spectral peaks used to generate molecular images is a critical step in all imaging mass spectrometry strategies. High mass resolution measurements as well as accurate mass can be utilized for compound assignment. Often other strategies need to be combined, such as using MALDI-tandem mass spectrometry or alternate analytical strategies to identify the types of compounds present on the surface.
MALDI-Tandem Mass Spectrometry
Tandem mass spectrometry is a logical choice for identifying the different compound ions generated from the surface. Limiting factors are the resolution of precursor ion selection, fragmentation efficiency, and method sensitivity. Often only a few mass spectrometry experiments can be performed at the same location. The number of experiments that can be conducted at a single location still depends on the number of laser shots that provide signal. Interleaved imaging scans performing tandem experiments at adjacent locations can partially overcome this issue. Once fragmentation patterns are known, multiple reaction monitoring can be applied to determine compound distributions.
LC-MS/MS Identification
Studies can use complementary tissue homogenization and extraction to generate a library of tissue components. LC-MALDI can also be utilized to address mixture complexity, increase sensitivity, and lower ion suppression effects. Comparative analysis of MALDI spectra observed in direct MALDI imaging experiments can be used as part of an identification strategy. In these studies, tandem MS can be used to identify individual compound components found on the LC-MALDI targets.
Transferred from Guangzhou Weiping Technology Service Co. Original link:https://lab.vogel.com.cn/c/2021-01-15/1078509.shtml